By Niklas Krausse
It is early spring 2020. What has started as a local outbreak in China has by now paralyzed many countries in Europe and the rest of the world. In one way or another, most of us have been affected by the recent coronavirus disease (COVID-19) pandemic. There is still a lot unknown about the pathogen itself, and despite tremendous efforts from scientists all over the world, a safe and effective vaccine is still some way off. But perhaps a look back into the past could provide some insights into how viruses from the same group infect cells and which intervention strategies are suggested. After all, this is not the first encounter humans have experienced with coronaviruses.
It is early spring 2020. What has started as a local outbreak in China has by now paralyzed many countries in Europe and the rest of the world. In one way or another, most of us have been affected by the recent coronavirus disease (COVID-19) pandemic. There is still a lot unknown about the pathogen itself, and despite tremendous efforts from scientists all over the world, a safe and effective vaccine is still some way off. But perhaps a look back into the past could provide some insights into how viruses from the same group infect cells and which intervention strategies are suggested. After all, this is not the first encounter humans have experienced with coronaviruses.
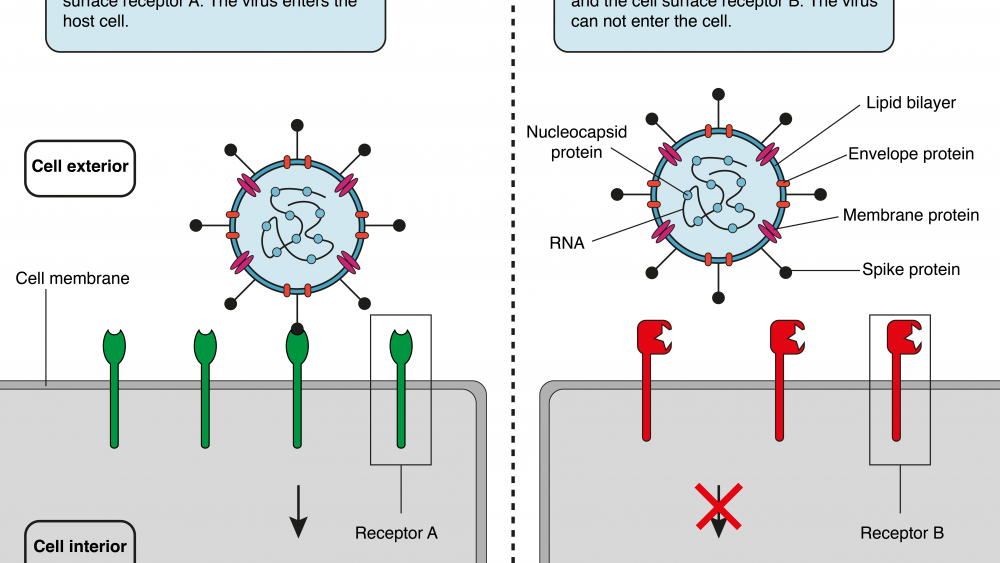
In 2002, an outbreak of severe acute respiratory syndrome (SARS) was reported. A virus that was designated SARS-CoV had crossed the species-boundaries (a process called zoonosis) and was identified as the cause of the disease. The pathogen responsible for the current outbreak (SARS-CoV-2) belongs to the same group of viruses as SARS-CoV, which are called Coronaviridae. In general, members of the Coronaviridae family consist of three structural components (proteins): spike (S), envelope (E), and membrane (M). These three proteins make up the outer structure of the virus, the so-called virus envelope. The genetic material (single stranded RNA in this case) is wrapped around the nucleocapsid (N) protein and located in the virus envelope. All viruses have to find a way to infiltrate their target cells in order to make use of their host’s internal protein-making machineries (ribosomes) to produce more virus particles. Understanding exactly how the virus enters the host provides vital information regarding the virus life cycle, and may help to stop the progression of the virus.
In 2003, Li and colleagues addressed this question in their work “Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus”, which they published in the prestigious scientific journal Nature.1 Previous studies had identified the S protein as the part of the virus that interacts with its target cells, similarly to a key in a lock. In order to identify the corresponding counterpart of the S protein on the host cell, they created a fusion protein with parts from the viral S protein and parts from human antibodies (important for detection of the fusion protein later in the study). Replicating the “key” of the virus, they managed to capture the “lock” on cells accessible to the virus. The “lock” turned out to be Angiotensin-converting enzyme 2 (ACE2). To further prove that the correct receptor had been identified, they artificially expressed ACE2 on cells that usually do not express it. And voilà – the fusion protein recognized the cells that were previously invisible to the virus again. Interestingly, they also found that adding a soluble version of the ACE2 receptor to cells vulnerable to viral infection prevented virus uptake by intercepting the virus. Blocking the ACE2 receptor with an antibody also hindered the virus from entering the cells. These effects were specific to ACE2, since the same experiments were performed with the closely related ACE1 receptor and did not prevent virus entry. In their work, they identified ACE2 as the functional receptor for SARS-CoV. Importantly, it has been shown before that ACE2 is expressed on a variety of different tissues in humans including the lungs, the heart, the kidneys and the gastrointestinal tract.
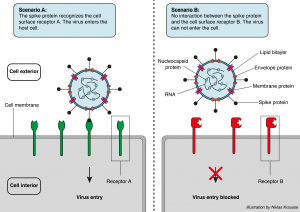
Seventeen years later, Hoffmann and colleagues were wondering if the newly emerged SARS-CoV-2 virus uses the same entry route as SARS-CoV.2 The S-proteins of both viruses are approximately 76% identical, and both viruses seem to affect the same spectrum of cell lines. Besides many structural similarities, ACE2 was indeed used by SARS-CoV-2 to enter the host cells. They performed the same trick as Li et al. and artificially expressed ACE2 on cells that then became susceptible to infection by SARS-CoV-2. Furthermore, they showed that virus entry depends on yet another protein located on the host cell. The enzyme, with the fancy name TMPRSS2, primes (or activates) the S-protein of the virus. Upon activation, the S-protein is cleaved into two subunits. Subunit 1 of the cleaved S-protein interacts with the target receptor on the host cell (ACE2) and establishes the first contact, almost like a molecular handshake between the virus and the cell. Subunit 2 of the cleaved S-protein then fuses the viral and host membrane together and completes the entry.3 One possible treatment approach would be to block the activity of the priming enzyme, and Hoffmann et al. have already shown that the clinically proven inhibitor “camostat mesylate” partially blocks S-protein driven entry of SARS-CoV-2.
This is just one example how research conducted almost 20 years earlier on a related virus has helped to accelerate the efforts made in understanding the biology behind a present threat. If you are interested to find out more about past and present vaccination approaches against SARS-CoV and SARS-CoV-2 stay tuned for our next blog post!